|
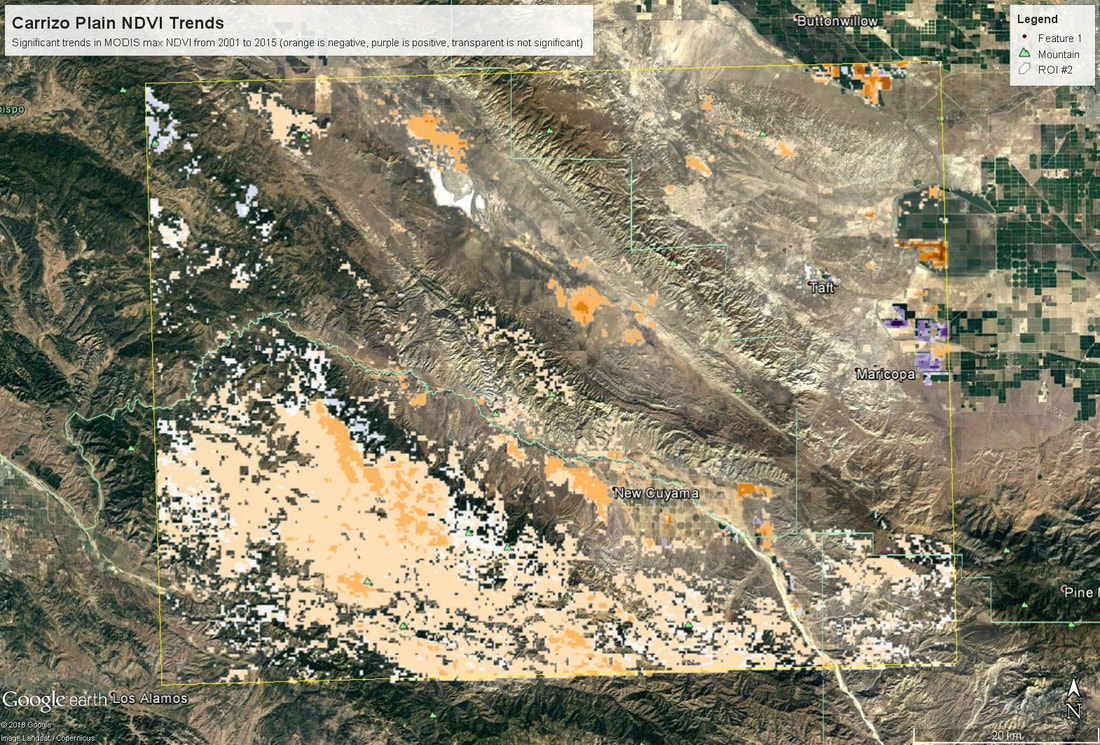
Last week and on Monday of this week we continued working in R, this time using actual remotely sensed data. This is a continuation of blog posts for days 1 (link) and day 2 (link) - I didn't include a post for day 3 because it was mostly catching up on the goals from day 2. Students have been doing awesome, however, so now we are attempting a longer set of code and introducing a bunch of new skills. The goals are (1) see how to download polygon data from AppEEARS; (2) open up a netCDF file in R (an unfamiliar file format for most of the students); (3) rasterize and summarize the MODIS data (to here on day 4); (4) write functions to map trend slopes and their significance; and (5) export that data as a KML so we can look at it in GoogleEarth (day 5). The code is posted here and students were given a printed out version of the 'workflow' (including instructions on how to access AppEEARS). Next time I teach this course I might have the students request their own data in advance, it just takes too long to get it back in time to request the data and work with it in the same session (my first request took about 30 min to be ready). All of this code should run on a different netCDF file, though, so they can do that on their own time if they want. Here's the short 'accessing AppEEARS' writeup I provide (and demonstrate in class):
Overall this went pretty smoothly. Since it ran in to day 5, some students were able to fold in their own data, which was cool to see. The only major hiccup was learning that if you create a KML in R using the raster::KML function, it generates both a .kml and a .png file (I think the png holds the color info?) - and in order to look at this in Google Earth, you need both of those files to be in the same folder.
My hope is that this exercise will serve as a starting off point for students to think about other things they could map with a time series of data. What about the variance in each pixel? trends in annual minima? One thing I'm curious to try is using trend decomposition across an image (stats::stl in R), though I'm not sure how I would summarize that info to make a map that was meaningful... but it could be cool! |
ERSAM Lab ThoughtsThis is where we occasionally post things that are not research, but more than 280 characters. If you want to know what we're working on now, this would be a good place to look. Code will be linked to here and posted on github. Archives
November 2018
Categories |

 RSS Feed
RSS Feed